
By now, most of us have read the stories about the upheaval DNA testing can cause by exposing unwanted pregnancies, extramarital affairs and secret adoptions.
What is often less appreciated is how it is upending scholars’ understanding of human history and evolution, including when homo sapiens migrated from Africa to Europe and Asia and whether they interbred with now extinct hominids when they got there. What they’ve learned has helped bolster some long-held theories, discredit others and — in a few instances — bust some urban myths.
That’s what happened in 2016, when a group of researchers at Yale University dusted off some old jars at the root of a boast by The Explorers Club that it had served mammoth at its annual dinner in New York City in 1951.
Members of the club had identified the meat as having come from frozen remains found by some of its members on Akutan Island off Alaska, according to media reports at the time. However, when a member later donated a sample of the meat to a museum, he said it had been taken from the remains of Megatherium, an extinct giant ground sloth thought to have lived only in South America.
The sample eventually ended up at the Yale Peabody Museum of Natural History. Even though the tissue had been stored in isopropyl alcohol or ethanol for decades, researchers in 2015 used modern lab techniques to extract enough DNA from the tissue to show it came from the green sea turtle (Chelonia mydas) – likely the same one served in a soup on the night in question according to a menu preserved from the famous banquet. [1]
The DNA data explosion
While scientists had long doubted the veracity of the mammoth meat myth, the finding illustrates how a surge of innovation since the completion of the Human Genome Project in 2003 has influenced decades-old debates in archeology, anthropology, linguistics, paleontology and other fields beyond medicine.
One of the biggest innovations has come in the area of high-throughput sequencing, which layers massively parallel microprocessing on top of the Sanger sequencing technology used for decades to scale up the search for tell-tale variations in the 3.2 billion base-pair human genome. Also referred to as “next-generation sequencing,” or NGS, the technology uses specially designed chips to simultaneously scan millions of strands of DNA and sequence hundreds to thousands of genes at a time.
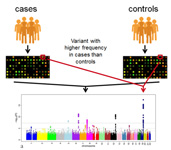 NGS gave rise to genome wide association studies (GWAS), which medical researchers use to look for patterns across large swaths of DNA taken from thousands of individuals in search of single-nucleotide polymorphisms (SNPs) and other genetic variations that might elucidate the causes of cancer and other diseases.
NGS gave rise to genome wide association studies (GWAS), which medical researchers use to look for patterns across large swaths of DNA taken from thousands of individuals in search of single-nucleotide polymorphisms (SNPs) and other genetic variations that might elucidate the causes of cancer and other diseases.
By revealing how some SNPs appear more frequently in some populations than others, GWASs have led to the development of “ancestry informative markers” (AIMs). Genetic anthropologist are using the growing amount of DNA recovered from ancient human remains to create datasets of AIMS that can be used to estimate the makeup of prehistoric populations with greater geographic and chronological precision in less time and at lower cost. Today, a scientist can download sequencing data on up to 1.23 million segments of DNA from thousands of ancient and modern human genomes simply by logging into a website maintained by the David Reich Lab at Harvard Medical School for free.[2]
The number of human genomes stored by direct-to-consumer DNA testing services, meanwhile, is on pace to surpass 100 million by early 2021, according to an article published by The MIT Technology Review earlier this year. [3] That would mark a four-fold increase in just three years. The growth will increasingly be fueled by demand for clinical-grade tests consumers can use to manage their health.
The cost of same-day, clinical-grade sequencing of entire human genomes is fast approaching the sub-$1,000 price point deemed critical to mass adoption of precision medicine. The Broad Institute, which sequenced its 100,000th human genome in 2018, forecast that 13.5 million whole human genomes will be sequenced for medical purposes by 2023, up from 1.5 million at the end of 2018. [4]
Given the dominance of Ancestry and 23andMe in both genealogical and medical testing, it’s likely innovation will flow between these sectors much more quickly in the decade ahead.
Unfurling our genetic maps
Thirty years ago, scientists used genomics to trace human migration across continents. Today, they can reveal migration patterns within regions — sometimes even within a single country over the period of a few generations.
In 2008, for instance, researchers who analyzed genetic variation at 200,000 locations, or loci, on the DNA of 3,000 Europeans concluded that an individual’s DNA could “be used to infer their geographic origin within a few hundred kilometres.[5]
“Even more surprising, when a new European person’s genome was analyzed, the researchers could predict where that person was from within a few hundred kilometers,” the authors wrote.
Two years later, an analysis of DNA chip data showed the median proportion of European ancestry among 365 African Americans sampled was 18.5 percent. [6]
In 2012, AncestryDNA announced it had developed a DNA test that could analyze a person’s genome at more than 700,000 locations and cross reference a worldwide DNA database to provide insights that might help them find descendants as far back as the mid-1700s. This meant customers of West African descent could narrow down their ancestry to one of six smaller regions or countries. Similarly, those who had previously been told they had ancestors in Great Britain could learn whether they had lived in England or Ireland and descendants with Southern European heritage could learn whether their clan came from Italy, Greece or the Iberian Peninsula.
 In a study published four years later, Ancestry researchers would explain how NGS and sophisticated computer modeling enabled them to glean hard-to-find genetic variations called “identity-by-descent” (IBD) markers from DNA to estimate “fine-scale population structure” in the United States in the years following the Revolutionary war by. [7]
In a study published four years later, Ancestry researchers would explain how NGS and sophisticated computer modeling enabled them to glean hard-to-find genetic variations called “identity-by-descent” (IBD) markers from DNA to estimate “fine-scale population structure” in the United States in the years following the Revolutionary war by. [7]
Researchers had long relied on modeling changing variations in allele frequencies in mitochondrial DNA to estimate the structure of past populations because they are easier to find. But such alleles, which are inherited only from the mother, can take centuries to change, making them useless for finding common ancestry in the New World since the Spanish Conquest.
IBD markers are long segments of autosomal DNA held in common by Individuals because they share a recent common ancestor. The problem is they are extremely hard to find because our autosomal DNA is formed by combining and randomly reshuffling the DNA inherited from each parent. It is located on 22 of the 23 human chromosomes that account for 3.1 billion of the 3.2 billion base pairs that make up our DNA.
In their study, Ancestry’s researcher noted that the chances of a segment of autosomal DNA in two or more individuals being identical due to a common ancestor dating back four or more generations is less than 1 percent. Ancestry researchers, however, were able to able to find 500 million IBD markers among 709,358 SNPs they identified thanks to the sheer size of their DNA database.
“After years of hard work, and a lot of rigorous statistics, we developed a novel scientific methodology that looks at how specific groups of people are connected through their DNA, what places they called home, and which migration paths they followed to get there – allowing genetics to reveal the history in a more recent time period than ever before,” AncestryDNA crowed on its blog.
Rewriting prehistory
Anthropologists have made great strides extracting, purifying and amplifying aDNA to shed new light on human evolution and migration going back tens and even hundreds of thousands of years.
 Swedish molecular geneticist Svante Pääbo described several of these advances in his 2014 memoir “Neanderthal Man,” which recounted the painstaking steps his team took to overcome contamination of Neanderthal bones and DNA by archeologists, museum curators and lab technicians.
Swedish molecular geneticist Svante Pääbo described several of these advances in his 2014 memoir “Neanderthal Man,” which recounted the painstaking steps his team took to overcome contamination of Neanderthal bones and DNA by archeologists, museum curators and lab technicians.
The team was ultimately able to extract, amplify, copy and sequence enough aDNA from the remains of three Neanderthals recovered decades earlier to show that about two percent of modern Europeans’ DNA comes from Neanderthals. In a landmark study published in 2010, the team estimated as much as 4.8 percent of the DNA of some modern Southeast Asian populations comes from another extinct branch of hominids called Denisovans. [8]
Neanderthals first appear in the fossil record about 800,000 years ago and are thought to have co-existed with homo sapiens in Eurasia from about 80,000-30,000 years ago, when Neanderthals appear to have gone extinct.
To the surprise of Pääbo and many of his colleagues at the Max Planck Institute for Evolutionary Anthropology, the frequency of certain SNPs indicated that homo sapiens interbred with both extinct hominids after migrating out of Africa.
Subsequent research has found traces of Denisovan, rather than Neanderthal DNA, in some Native American populations. It is now believed that aDNA accounts for as much as 40 percent of the variation in the human genome.
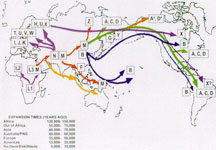
In 2012, National Geographic began selling consumers a DNA test kit capable of tracing both maternal and paternal ancestry back tens of thousands of years in what essentially amounted to a global DNA crowdsourcing campaign. The Geno 2.0 Kit, which was developed under the aegis of Lewontin protégé Dr. Spencer Wells at the University of Texas at Austin, used NGS to analyze 300,000 genetic markers as part of The Genographic Project. National Geographic had launched the project with IBM and other partners in 2005 in a bid in a bid to uncover the story of how our ancestors migrated out of Africa and spread to Europe and Asia. [9]
To ensure North Americans and Europeans did not skew the results, project sponsors set up labs overseas for local scientists to collect and analyze DNA samples. By the time the project ended in 2018, 1 million kits had been sold in 140 countries.
In 2013, Pääbo was surprised again when a team he led found mitochondrial DNA extracted from a 430,000-year-old human femur excavated at Sima de los Huesos was more akin to Denisovan than Neanderthal DNA. Further DNA investigation may reveal the bones belonged to a yet-to-be identified ancestor of both hominids. In the meantime, however, many scientists are wondering if Denisovans emerged hundreds of thousands of years earlier and on an entirely different continent than once believed.[10]
A year later, a comparison of DNA extracted from a 7,000-year-old farmer found in Germany and eight, 8,000-year-old hunter-gatherers from Luxembourg and Sweden with DNA from other ancient and modern humans indicated Near Eastern migrants played a major role in introducing agriculture to Europe.[11]
Since the beginning of 2019, aDNA studies have upset two widely held notions about migration patterns in the Old World and shed light on one of the most fascinating mysteries of modern Europe.
 In one instance, an analysis of aDNA from 524 previously unsampled individuals from Iran, south-central Asia and northern Pakistan undermined the so-called “Anatolian hypothesis,” which holds that farmers from what is now Turkey introduced agriculture and Indo-European languages to Southern Asia. [12]
In one instance, an analysis of aDNA from 524 previously unsampled individuals from Iran, south-central Asia and northern Pakistan undermined the so-called “Anatolian hypothesis,” which holds that farmers from what is now Turkey introduced agriculture and Indo-European languages to Southern Asia. [12]
Another study suggests people were moving into Iberia from Africa more than 3,000 years before the rise of the Roman Empire.[13] The research is based on an analysis of DNA from 403 ancient Iberians dated between 6000 BC and 1600 AD, 975 ancient people from outside Iberia, and 2,862 present-day people. The findings could help explain why the mysterious Basques of Northern Spain are the only population in Western Europe today that does not speak an Indo-European language.
References:
[1] https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0146825
[2] https://reich.hms.harvard.edu/downloadable-genotypes-present-day-and-ancient-dna-data-compiled-published-papers
[3] https://www.technologyreview.com/s/612880/more-than-26-million-people-have-taken-an-at-home-ancestry-test/
[4] https://www.broadinstitute.org/news/broad-institute-sequences-its-100000th-whole-human-genome-national-dna-day
[5] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2735096/
[6] https://www.pnas.org/content/107/2/786/
[7] http://www.nature.com/articles/ncomms14238
[8] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5100745/
[9] https://genographic.nationalgeographic.com/about/
[10] https://www.nature.com/news/hominin-dna-baffles-experts-1.14294
[11] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4170574/
[12] https://science.sciencemag.org/content/365/6457/eaat7487
[13] https://www.nytimes.com/2019/03/14/science/iberia-prehistory-dna.html

Charlie Lunan is a science writer with a background in financial journalism who follows the intersection of genetics, evolution, infectious disease, pollution and business. In addition to his freelance work, he produces a website promoting outdoor recreation and sustainable living within a four-hour drive of his home in Charlotte, N.C.
